PIPUNCULIDAE (DIPTERA) FROM THE CALDERA DE TABURIENTE NATIONAL PARK, LA PALMA (CANARY ISLANDS, SPAIN) — INVESTIGATING THE
MORPHOLOGICAL AND MOLECULAR VARIABILITY IN A NEW SPECIES OF BIG-HEADED FLIES
Christian Kehlmaier1,* & Miguel Ángel Alonso-Zarazaga2
1Senckenberg Natural History Collections Dresden, Museum of Zoology, Königsbrücker Landstrasse 159, D–01199 Dresden, Germany. ORCID iD: https://orcid.org/0000-0001-9622-0566 E-mail: kehlmaier@web.de
2Museo Nacional de Ciencias Naturales, Departamento de Biodiversidad y Biología Evolutiva, C/. José Gutiérrez Abascal 2, E-28006
Madrid, Spain. ORCID iD: https://orcid.org/0000-0002-6991-0980 E-mail: zarazaga@mncn.csic.es
*Corresponding author: kehlmaier@web.de
| |
Abstract
The present paper is a result of the project “Inventory and study of the invertebrate fauna of the Caldera de Taburiente National
Park on La Palma, Canary Islands. Among the four species of Pipunculidae recorded, Chalarus guanche Kehlmaier sp. nov. is described and also recorded from Madeira, whereas Tomosvaryella freidbergi De Meyer, 1995 and T. parakuthyi De Meyer, 1995 are first records for La Palma. The morphological and molecular variability of C. guanche sp. nov. is studied and the presence of intragenomic variation in ITS2 rDNA is discussed.
http://zoobank.org/urn:lsid:zoobank.org:pub:D6A9AA88-A717-4AC2-AA8C-B32F1C91081E
Key words: Diptera; Pipunculidae; Chalarus guanche sp. nov.; Caldera de Taburiente National Park; La Palma; Canary Islands; new species; DNA-barcoding; intragenomic variation; COI; ITS2.
|
| |
RESUMEN
Pipunculidae (Diptera) del Parque Nacional de la Caldera de Taburiente, La Palma (Islas Canarias, España) — Investigando la
variabilidad morfológica y molecular de una nueva especie de moscas cabezonas
Este trabajo es el resultado del proyecto “Inventario y estudio de la fauna invertebrada del Parque Nacional de la Caldera
de Taburiente” en la isla de La Palma, Islas Canarias. De las cuatro especies recogidas, se describe Chalarus guanche sp. nov. que se registra asimismo de Madeira, y Tomosvaryella freidbergi De Meyer, 1995 y T. parakuthyi De Meyer, 1995 son nuevos registros para La Palma. Se estudia la variabilidad morfológica y molecular de C. guanche sp. nov. y se discute la presencia de variación intragenómica en el ADNr ITS2.
Palabras clave: Diptera; Pipunculidae; Chalarus guanche sp. nov.; Parque Nacional de Caldera de Taburiente; La Palma; Islas Canarias; nueva especie; DNA-barcoding; variabilidad intragenómica; COI; ITS2.
|
IntroductionTOP
Slightly more than 1.400 species of Pipunculidae are known from all around the world (Rafael & Skevington, 2010). Their common name ‘big-headed flies’ refers to the globular head that is almost entirely covered by their large compound
eyes. This panorama view enables them to precisely navigate in dense vegetation, and helps the females to detect suitable
hosts for their endoparasitic larvae, which develop mainly within larval and adult Auchenorrhyncha, but have specialised on
adult Tipulidae in Nephrocerus Zetterstedt, 1838 (see Rafael & Skevington, 2010 for a brief review of the family’s biology).
De Meyer et al. (2001) present a faunistic overview of the pipunculid fauna known from the Macaronesian archipelagos of Canary Islands (Spain) and
Madeira (Portugal), listing 13 species for the Canary Islands, including five for La Palma, and three species known from Madeira.
With the current paper, we complement this listing by studying material mainly resulting from a two year trapping survey conducted
in the Caldera de Taburiente National Park on La Palma (Domingo-Quero et al., 2003).
Material and methodsTOP
The material studied originates from the project “Inventory and study of the invertebrate fauna of the Caldera de Taburiente
National Park”. The trapping setup covered six sites (Table 1), ranging from 750–2,345 m above sea level, each equipped with a Malaise trap (MT) and a yellow pan trap (YT) placed under
the middle wall of the Malaise trap. The traps were emptied at intervals between 4 and 15 days (average of 7.75 days) from
August 1999 to July 2001 by T. Domingo-Quero and A. Sánchez-Ruiz. For further details see Domingo-Quero et al. (2003). The material is currently ethanol preserved and in the collections of the Museo Nacional de Ciencias Naturales in Madrid
(MNCN), Senckenberg Deutsches Entomologisches Institut in Müncheberg (SDEI) and Senckenberg Museum für Tierkunde Dresden (SMTD).
Table 1.— List of trapping localities in order of ascending altitudes.
Tabla 1.— Lista de localidades de trampeo por orden
de altitud ascendente.
| Locality |
Altitude |
UTM coordinates |
| Playa del Rio Taburiente |
750 m |
UTM 28RBS1980-1 |
| Barranco de Las Traves |
1,068 m |
UTM 28RBS1780-2 |
| Lomo de las Chozas |
1,299 m in 1999; 1,277 m in 2000 & 2001 |
UTM 28RBS2077-3 |
| Roque de la Cumbrecita |
1,377 m |
UTM 28RBS2177-1 |
| Roque de los Muchachos |
2,250 m |
UTM 28RBS2084-3 |
| Pico de la Cruz |
2,345 m |
UTM 28RBS2183-1 |
Morphological terminology follows Kehlmaier & Assmann (2008), including the following abbreviations: LW—length of wing; MWW—maximum width of wing; LS—length of pterostigma; LSC—length
of second costal section of wing; LTC—length of third costal section of wing; LFC—length of fourth costal section of wing;
psr—posterior setal row of front femur; pvsr—posteroventral setal row of mid femur; aasr—anterior/anterodorsal setal row of
hind femur; pdsr—posterodorsal/dorsal setal row of hind femur; LT35—maximum length of tergites 3–5; WT2—maximum width of tergite
2; Lmtdp—length of membranous tip of the distiphallus; Ltdp—length of tip of distiphallus.
Morphometric measurements and resulting ratios were investigated in a series of up to 25 ethanol preserved females. The following
aspects were investigated: diameter of largest frontal facet (DFF) (n=25); width of frons at its narrowest point (FN) (n=23);
width of frons at level of anterior (median) ocellus (FMO) (n=25); FN:FMO (n=23); DFF:FN (n=23); length of tergite 9 (piercer)
in lateral view (n=24); wing length in dorsal view (n=25).
Molecular work was carried out at MNCN and SMTD according to well established standard procedures outlined in Kehlmaier & Assmann (2010). A fragment of the 5’ end of the mitochondrial coding gene cytochrome oxidase subunit I (COI) was obtained using the primer
pair LCO1490-JJ (5’–CHACWAAYCATAAAGATATYGG–3’) and HCO2198-JJ (5’–AWACTTCVGGRTGVCCAAARAATCA–3’) (Astrin & Stüben, 2008), whereas part of the nuclear nontranscribed ribosomal Internal Transcribed Spacer Region 2 (ITS2) was generated with the
primer pair ITS2A (5’–TGTGAACTGCAGGACACAT–3’) and ITS2B (5’–TATGCTTAAATTCAGGGGGT–3’) (Beebe & Saul, 1995). The resulting sequences were checked by eye for base-calling errors and manually aligned with the software BioEdit (Hall, 1999). Molecular analyses were computed with MEGA6 (Tamura et al., 2013), resulting in unrooted neighbour-joining (NJ) trees of uncorrected pairwise genetic distance (p-dist), using all sites and
pairwise deletion for missing data. The sequence data is deposited at the European Nucleotide Archive (ENA) under LT715904–38
(COI) and LT715939–81 (ITS2), the individual accession numbers being listed in Appendix 1.
ResultsTOP
Systematic account of big headed flies of La PalmaTOP
The 78 specimens collected at the Caldera de Taburiente National Park comprise four species. With 62 individuals, the genus
Chalarus was dominant. All trapping localities yielded specimens except “Pico de la Cruz”.
PIPUNCULINAETOP
Dasydorylas setosus (Becker, 1908)
MATERIAL (MT: 2♂♂ 5♀♀; YT: 3♀♀). SPAIN, Canary Islands, La Palma, Caldera de Taburiente National Park: Playa del Rio Taburiente: 15.V.2000,
1♀ (MNCN_Ent 202044, YT), coll. MNCN; 30.V.2000, 1♂ (MT), coll. SMTD; 19.VI.2000, 1♀ (MNCN_Ent 202045, MT), coll. MNCN.— Barranco
de las Traves: 5.VI.2000, 1♀ (MNCN_Ent 202042, YT), coll. MNCN; 19.VI.2000, 1♀ (YT), coll. SMTD; 27.VI.2000, 1♂ (MNCN_Ent
202043, MT), coll. MNCN.— Lomo de las Chozas: 1.VI.2000, 1♀ (MNCN_Ent 202047, MT), coll. MNCN.— Roque de la Cumbrecita: 1.VI.2000,
1♀ (MNCN_Ent 202041, MT), coll. MNCN; 21.VI.2000, 1♀ (MNCN_Ent 202046, MT), coll. MNCN.— Roque de los Muchachos: 20.VI.2001,
1♀ (MT), coll. SMTD.
REMARKS. Dasydorylas setosus was originally described from specimens collected on Gran Canaria, La Palma and Tenerife, and was redescribed by Kehlmaier (2005). Its current distribution comprises Canary Islands (La Gomera, El Hierro, La Palma, Tenerife, Gran Canaria), mainland Spain,
Madeira and Morocco. Here, the species was recorded from 15th May to 27th June. Literature records indicate that it can be
found all year round on Canary Islands.
Tomosvaryella freidbergi De Meyer, 1995
MATERIAL (MT: 2♀♀). SPAIN, Canary Islands, La Palma, Caldera de Taburiente National Park: Roque de la Cumbrecita: 1.VI.2000, 1♀ (MNCN_Ent
202050, MT), coll. MNCN.— Roque de los Muchachos: 4.VII.2001, 1♀ (MT), coll. SMTD.
REMARKS. Representing the first record for La Palma, T. freidbergi was previously recorded from Canary Islands (Fuerteventura, La Gomera, Tenerife) by De Meyer et al. (2001). For species recognition see De Meyer (1995) and Földvári & De Meyer (1999). Tomosvaryella freidbergi has been recorded from Canary Islands, mainland Portugal, mainland Spain, mainland France, Israel, Hungary, Egypt, Iran,
Kazakhstan and Kyrgyz Republic. The material at hand was collected from 1st June to 4th July. Literature records for Canary
Islands range from February to July and September.
Tomosvaryella parakuthyi De Meyer, 1995
MATERIAL (MT: 2♀♀; YT: 2♂♂). SPAIN, Canary Islands, La Palma, Caldera de Taburiente National Park: Barranco de las Traves: 19.VI.2000,
1♂ (MNCN_Ent 202048, YT), coll. MNCN.— Roque de los Muchachos: 31.V.2001, 1♀ (MNCN_Ent 202049, MT), coll. MNCN; 15.VII.2001,
1♀ (MT), coll. SMTD; 17.VII.2001, 1♂ (YT), coll. SMTD.
REMARKS. First record for La Palma, yet previously cited from Gran Canaria by De Meyer et al. (2001). The present material has been recorded between 31st May and 17th July. The specimen from Gran Canaria was collected at the
end of February. So far, T. parakuthyi is known from Canary Islands, Egypt, Iran and Israel.
CHALARINAETOP
Chalarus guanche Kehlmaier sp. nov.
Chalarus perplexus in De Meyer et al. (2001)
Chalarus sp. near zyginae in Kehlmaier & Assmann (2008)
http://zoobank.org/urn:lsid:zoobank.org:act:7FAE7CE3-4DCA-4425-81AF-EC870CDDBDEE
Figs. 1–3
|
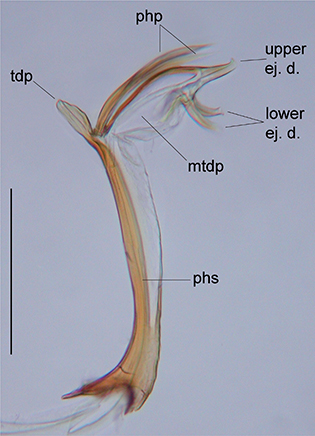 Fig. 1.— Male genitalia of C. guanche sp. nov. in lateral view. Abbreviations: lower ej.d., lower ejaculatory ductuli; mtdp, membranous tip of distiphallus; php, phallic
processes; phs, phallic shaft; tdp, tip of distiphallus; upper ej.d., upper ejaculatory duct. Scale bar: 0.1 mm. Fig. 1.— Male genitalia of C. guanche sp. nov. in lateral view. Abbreviations: lower ej.d., lower ejaculatory ductuli; mtdp, membranous tip of distiphallus; php, phallic
processes; phs, phallic shaft; tdp, tip of distiphallus; upper ej.d., upper ejaculatory duct. Scale bar: 0.1 mm.
Fig. 1.—
Genitalia masculina de C. guanche sp. nov. en vista lateral. Abreviaturas: lower ej. d., túbulos eyaculadores inferiores; mtdp, ápice membranoso del distifalo; php,
procesos fálicos; phs, asta fálica; tdp, ápice del distifalo; upper ej.d., tubo eyaculador superior. Escala: 0,1 mm.
|
|
|
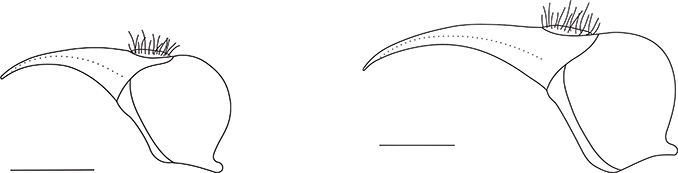 Figs. 2–3.— Female terminalia of C. guanche sp. nov. Scale bars: 0.1 mm. Fig. 2: Representative of clade A (no. 18). Fig. 3: Representative of clade B (no. 8). Figs. 2–3.— Female terminalia of C. guanche sp. nov. Scale bars: 0.1 mm. Fig. 2: Representative of clade A (no. 18). Fig. 3: Representative of clade B (no. 8).
Figs. 2–3.— Terminalia femenina de C. guanche sp. nov. Escalas: 0,1 mm. Fig. 2: Representante del clado A (nº 18). Fig. 3: Representante del clado B (nº 8).
|
|
HOLOTYPE. 13.VI.2000, ♂ (#29, YT), SPAIN, Canary Islands, La Palma, Caldera de Taburiente National Park, Barranco de las Traves, leg.
T. Domingo-Quero, coll. MNCN (MNCN_Ent 160689).
PARATYPES. SPAIN, Canary Islands, La Palma, Caldera de Taburiente National Park: Playa del Rio Taburiente: 6.III.2000, 1♀ (#25, MT).—
Barranco de las Traves: 22.VIII.2000, 1♀ (#2, MNCN_Ent 160706, MT).— Lomo de las Chozas: 8.III.2000, 1♀ (#21, MNCN_Ent 160705,
MT); 22.III.2000, 1♂ (#48, MNCN_Ent 160690, YT); 26.IV.2000, 1♀ (#15, MNCN_Ent 160702, MT); 21.VI.2000, 5♂♂ (#43–47
[1]
, MT), 1♀ (#14, MNCN_Ent 160695, MT); 6.VII.2000, 2♀♀ (#19–20[2]
, MT); 19.VII.2000, 3♂♂ (#40–42[3]
, MT); 4♀♀ (#10–13[4]
, MT); 26.VII.2000, 1♀ (#17, MT); 16.VIII.2000, 2♂♂ (#38–39, MT); 6.IX.2000, 1♀ (#18, MT).— Roque de los Muchachos: 25.VIII.2000,
1♀ (#27, MNCN_Ent 160707, MT). Paratypes in coll. SMTD: #17, #18, #25, #38, #39, #42, #43. Others in coll. MNCN.
NON-TYPE MATERIAL. SPAIN, Canary Islands, La Palma, Caldera de Taburiente National Park: Playa del Rio Taburiente: 17.IV.2000, 1♂ (#55, MT),
1♀ (#23, MNCN_Ent 202027, MT); 15.V.2000, 1♀ (#22, MT); 19.VI.2000, 1♂ (#57, MNCN_Ent 202029, YT); 3.VIII.2000, 4♂♂ (#58–61
[5]
, YT), 1♀ (#24, MNCN_Ent 202034, YT); 7.VIII.2000, 1♀ (#26, MNCN_Ent 202028, MT); 18.X.1999, 1♂ (#56, MT).— Barranco de las
Traves: 8.V.2000, 1♀ (#8, MT); 30.V.2000, 1♀ (#6, MNCN_Ent 160708, MT); 5.VI.2000, 1♂ (#35, MT); 13.VI.2000, 1♂ (#30, MNCN_Ent
202018, MT); 19.VI.2000, 1♀ (#1, MNCN_Ent 202019, MT); 27.VI.2000, 1♂ (#33, MNCN_Ent 202020, MT); 4.VII.2000, 1♂ (#34, MNCN_Ent
202021, MT); 10.VII.2000, 1♂ (#28, MNCN_Ent 202023, YT); 24.VII.2000, 1♂ (#32, YT), 1♀ (#3, MNCN_Ent 202022, MT); 3.VIII.2000,
2♀♀ (#4, MNCN_Ent 202024, #5, MNCN_Ent 202025, YT); 7.VIII.2000, 1♂ (#36, MT), 1♀ (#7, MT); 18.IX.2000, 1♂ (#31, MNCN_Ent
202026, YT).— Lomo de las Chozas: 5.I.2000, 1♂ (#51, MNCN_Ent 202035, MT); 26.IV.2000, 1♂ (#49, MT), 1♀ (#16, MT); 1.VI.2000,
1♂ (#37, MNCN_Ent 202036, MT), 1♀ (#9, MT); 6.VII.2000, 3♂♂ (#52–54[6], MT); 26.VII.2000, 1♂ (#50, MNCN_Ent 202039, MT).— Roque de los Muchachos: 27.VII.2000, 1♂ (#62, MNCN_Ent 202040, YT). Specimens
in coll. SMTD: #7, #8, #22, #32, #35, #36, #49, #54, #55, #56. Others in coll. MNCN.— SPAIN, Canary Islands, La Palma, south
of Barlovento, laurisilva, 28°39’N 17°52’W, 800 m, 29.X.2002, 3♂♂ (DNA CK145, CK146, CK417), 1♀, leg. et coll. C. Kehlmaier.—
PORTUGAL, Madeira, path between Boca de Encumeada and Boca dos Corgos, S of Ribeiro do Póco, 1 km W of Fenda do Ferreiro,
moist steep slope, 1200–1250 m, 30.V.1987, 1♂ (M1, X572), leg. W. Barkemeyer, coll. SDEI.— PORTUGAL, Madeira, Fanal, laurel
on pasture, 12.IX.1986, 3♂♂ (M2–M4, L1583), leg. P. Ohm, coll. SDEI.— PORTUGAL, Madeira, Pico Facho, near Machico, herbaceous
vegetation, 300 m, 19.IX.1986, 1♂ (M5, L1578), leg. P. Ohm, coll. SDEI.
NOTE. The material at hand consists of 71 specimens. Sixty-two from the Caldera de Taburiente National Park (35♂♂, 24 MT, 11YT;
27♀♀, 24MT, 3YT)—unfortunately, specimens #9 and #16 were misplaced, and of specimens #37 and #49 only the genitalia remain—,
three males and one female from Barlovento (NE La Palma), and five male specimens from three localities on Madeira. Only specimens
attributed to clade A via ITS2 were included in the type series in order to prevent confusion if clade B or C might be considered
as a distinct species one day.
DIFFERENTIAL DIAGNOSIS. Chalarus guanche sp. nov. is part of the holosericeus species group as outlined in Kehlmaier & Assmann (2008, 2010). The taxonomic structure of this flock of closely allied taxa is still not satisfactorily resolved. Morphologically, females
are closest to C. zyginae Jervis, 1992 being slightly larger in size and having their frons not as strongly narrowed (at narrowest point ~1.5 times
largest frontal facet instead of 1.0), whereas males should be addressed as C. holosericeus agg. (aggregate species), as the genitalia of all taxa of this species group are virtually identical and cannot be used for
species separation except in the case of C. zyginae. Because of the cryptic nature of this species, only the holotype is described in detail, whereas the observed variability
of the females is characterised in Tables 2–3 and Figs 2–4.
Table 2.— Morphometric measurements and ratios in female C. guanche sp. nov. from La Palma.
Tabla 2.— Medidas morfométricas y proporciones en las hembras de C. guanche sp. nov. de La Palma.
| DFF: Diameter of largest frontal facet in mm (n=25). |
| 0.035 |
0.04 |
0.045 |
0.05 |
|
|
|
|
|
|
|
|
average |
median |
| 1 |
17 |
4 |
3 |
|
|
|
|
|
|
|
|
0.0418 |
0.04 |
| FN: Width of frons at its narrowest point in mm (n=23) — 2 specimens with distorted frons excluded (0.025 mm and 0.035 mm). |
| 0.04 |
0.045 |
0.05 |
0.055 |
|
|
|
|
|
|
|
|
average |
median |
| 4 |
7 |
11 |
1 |
|
|
|
|
|
|
|
|
0.0452 |
0.05 |
| FMO: Width of frons at level of anterior (median) ocellus in mm (n=25). |
| 0.07 |
0.09 |
0.095 |
0.1 |
|
|
|
|
|
|
|
|
average |
median |
| 1 |
15 |
6 |
3 |
|
|
|
|
|
|
|
|
0.0916 |
0.09 |
| FN:FMO (n=23). |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 0.42 |
0.44 |
0.47 |
0.50 |
0.53 |
0.56 |
0.57 |
0.58 |
|
|
|
|
average |
median |
| 1 |
2 |
3 |
7 |
1 |
7 |
1 |
1 |
|
|
|
|
0.51 |
0.50 |
| DFF:FN (n=23). |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 0.73 |
0.80 |
0.88 |
0.89 |
0.90 |
1.00 |
1.11 |
1.25 |
|
|
|
|
average |
median |
| 1 |
7 |
1 |
5 |
3 |
4 |
1 |
1 |
|
|
|
|
0.90 |
0.89 |
| Length of tergite 9 (piercer) in lateral view in mm (n=24)—1 specimen with lost ovipositor excluded. |
| 0.15 |
0.17 |
0.18 |
0.19 |
0.20 |
0.21 |
0.22 |
0.24 |
|
|
|
|
average |
median |
| 1 |
2 |
5 |
2 |
6 |
2 |
4 |
2 |
|
|
|
|
0.20 |
0.20 |
| Wing length in mm (n=25). |
| 1.5 |
1.55 |
1.7 |
1.75 |
1.8 |
1.85 |
1.9 |
1.95 |
2.05 |
2.1 |
2.15 |
2.25 |
average |
median |
| 1 |
2 |
1 |
3 |
2 |
5 |
1 |
2 |
1 |
4 |
1 |
2 |
1.89 |
1.85 |
Table 3.— Morphometric measurements and ratios in female C. guanche sp. nov. from La Palma corresponding to ITS2 clade A and B.
Tabla 3.— Medidas morfométricas y proporciones de las hembras
de C. guanche sp. nov. de La Palma correspondientes a los clados A y B del ITS2.
| |
♀♀ ITS2 clade A |
♀♀ ITS2 clade B |
| DFF (diameter of largest frontal facet) |
0.035–0.045 mm (n=14) |
0.04–0.05 mm (n=6) |
| FN (width of frons at narrowest point) |
0.04–0.05 mm (n=13) |
0.04–0.05 mm (n=5) |
| FMO (width of frons at level of anterior/median ocellus) |
0.07–0.95 mm (n=14) |
0.09–0.1 mm (n=6) |
| FN:FMO |
0.80–1.00 (n=13) |
0.90–1.25 (n=5) |
| DFF:FN |
0.44–0.57 (n=13) |
0.42–0.56 (n=5) |
| Tergite 9 |
0.15–0.21 mm (n=13) |
0.22–0.24 mm (n=7) |
| Wing length |
1.5–2.05 mm (n=14) |
2.1–2.25 mm (n=7) |
|
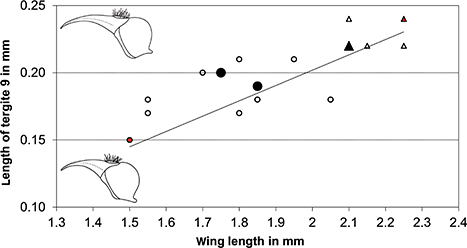 Fig. 4.— Relationship between wing length and length of tergite 9 in female C. guanche sp. nov. Circles represent clade A (n=13; small open circle: 1 specimen; large full circle: 2 specimens; small red circle: figured
specimen #18 (Fig. 2)); triangles represent clade B (n=7; small open triangle: 1 specimen; large full triangle: 3 specimens;
small red triangle: figured specimen #8 (Fig. 3)). Fig. 4.— Relationship between wing length and length of tergite 9 in female C. guanche sp. nov. Circles represent clade A (n=13; small open circle: 1 specimen; large full circle: 2 specimens; small red circle: figured
specimen #18 (Fig. 2)); triangles represent clade B (n=7; small open triangle: 1 specimen; large full triangle: 3 specimens;
small red triangle: figured specimen #8 (Fig. 3)).
Fig. 4.— Relación entre la longitud del ala y la del terguito 9 de las
hembras de C. guanche sp. nov. Los círculos representan el clado A (n=13; círculo pequeño vacío: 1 ejemplar; círculo grande relleno: 2 ejemplares; círculo
pequeño rojo: ejemplar figurado nº 18 (Fig. 2)); los triángulos representan el clado B (n=7; triángulo pequeño vacío: 1 ejemplar;
triángulo grande relleno: 3 ejemplares; triángulo pequeño rojo: ejemplar figurado nº 8 (Fig.3)).
|
|
DESCRIPTION OF HOLOTYPE. Male. Body length 1.9 mm. Head. Face black, silver-grey pollinose. Eyes separated, ommatidial facets slightly enlarged towards
the front. Frons black, silver-grey pollinose in lower quarter. At its narrowest point, width of 2 accompanying ommatidial
facets. Antenna dark brown. Pedicel with 2 short upper and 2 lower bristles, one of the latter longer than flagellum which
is of an ovoid-kidney shape and is slightly longer than wide. Vertex black. Ocellar triangle with 1 pair of long and 2 pairs
of short ocellar bristles. Occiput black, hardly visible in lateral view. Thorax. Entirely dark brown to black. Dorsal surface
of prescutum and scutum covered with rather widely spaced black setae, as in other species of the genus, the longest ones
towards the lateral and posterior margins (notopleural, supraalar and postalar bristles). Scutellum with 2 pairs of long black
marginal bristles, dorsally with 2 pairs of short setae. Wing and halter. Length: 1.9 mm. LW:MWW=2.8. Wing surface with a
weak brownish tinge and covered with microtrichia except near base. Pterostigma brown and incomplete (LS:LTC=0.8). LSC:LTC:LFC=6.8:5.5:1.0.
Wing venation incomplete, as in other members of Chalarus. Halter dark brown with stem narrowly white. Leg. Entirely dark brown except tarsal segments slightly paler (mid brown).
psr ~13 setae, mid brown; pvsr ~16 setae, light brown, apical one not extending beyond apex; aasr ~9 short setae, mid brown,
apical ones extending as far as apex; pdsr ~8 dark brown setae, apical ones extending as far as apex. Pulvilli shorter than
distitarsus. Abdomen. Gently ovate in dorsal view, LT35:WT2=1.1. Entirely dark brown. Setae mid to dark brown. Dorsally and
ventrally sparse and short, long and dense along lateral margins. Terminalia as in other members of the holosericeus species group (Fig. 1; see also figures in Kehlmaier & Assmann, 2008): surstyli symmetrical, with pronounced medial protuberances, best seen in dorsal view; gonopods symmetrical, broadened in
distal half; phallus with straight shaft; tip of distiphallus short and broad; Lmtdp:Ltdp slightly more than 3.0; phallic
processes symmetrical, slightly shorter than membranous tip of distiphallus and parallel to the latter; all three ejaculatory
ducts placed distally on membranous tip of distiphallus; ejaculatory apodeme parasol-shaped.
Female differs from male by the usual sexual dimorphisms. For morphometric measurements and ratios see Tables 2–3. Bristles of pedicel shorter; none longer than flagellum. Frontal facets moderately to greatly enlarged. Frons at narrowest
point about diameter of largest frontal facet. Frons with pairs of fronto-orbital setae. Tergites 2–5 laterally with shorter
setae. Ovipositor as in Figs. 2–3; one and a half to twice length of base and tergite 9 gently curved towards sternites. Base dark brown; piercer somewhat
paler towards apex.
ETYMOLOGY. The species is named after the native inhabitants of the Canary Islands. It is believed that the Guanches were of Berber
origin (North West African) (Fregel et al., 2009) and migrated to the Canary Islands around 1,000 BC or perhaps earlier.
DISTRIBUTION. The species has been recorded from La Palma (Canary Islands) and Madeira.
MORPHOLOGICAL VARIABILITY. The observed morphological variability in the material of Chalarus studied (Tables 2–3) raised the suspicion as to whether it actually contained several undescribed species. The degree of enlargement of the frontal
ommatidial facets (DFF) varies by ~40% (0.035–0.05 mm). Wing length, used to assess the size of the fly, by ~50% (1.5–2.25
mm). And the length of tergite 9 by ~60% (0.15–0.24 mm). In the latter, one can also observe that the longer tergite 9 is,
the stronger is its curvature (Figs. 2–3). When scoring length of tergite 9 against wing length, a positive correlation between these two morphological traits can
be observed (Fig. 4). However, although the extrema clearly differ, there does not seem to exist a precise division.
MOLECULAR ANALYSES. The Internal Transcribed Spacer Region 2 (ITS2) is a nontranscribed fragment of the ribosomal DNA (rDNA). Being nested between
the slow evolving 5.8S and 28S rDNA loci, ITS2 is renowned for its high evolutionary rate and has been successfully applied
for species delimitation and phylogenetic studies in Pipunculidae and other taxa (Kehlmaier & Assmann, 2010). The aligned ITS2 dataset for the Chalarus exiguus species group is 352bp long and consists of 51 specimens representing six species. In this study, the fragment had approximately
370bp and was sequenced in mostly excellent quality for 41 specimens. The NJ analysis of the ITS2 dataset, divides C. guanche sp. nov. into three clades, with two present on La Palma (clade A & B) and a third one on Madeira (clade C) (Fig. 5). With 0.6%, the uncorrected pairwise genetic distance is identical between all three clades (Table 4). Each clade has a single private (apomorphic) mutation. In addition, clade A has a 3bp long indel (Fig. 6). The minimum interspecific genetic distance between the species of the exiguus group ranges from 1.1% to 3.8% (Table 4). The 41 specimens fall into 26 clade A, 11 clade B and 4 clade C.
Table 4.— Uncorrected minimum interspecific genetic distance in % of Chalarus exiguus species group for ITS2 dataset.
Tabla 4.— Distancia genética interespecífica mínima no corregida en tanto por ciento del
grupo de especies de Chalarus exiguus para el conjunto de datos del ITS2.
| |
FM178160 |
FM178167 |
FM178169 |
FM178165 |
FM178171 |
clade A |
clade B |
clade C |
| C. holosericeus FM178160
|
— |
|
|
|
|
|
|
|
| C. exiguus FM178167
|
2.3 |
— |
|
|
|
|
|
|
| C. griseus FM178169
|
1.7 |
3.2 |
— |
|
|
|
|
|
| C. saxonicus FM178165
|
1.1 |
2.3 |
2.3 |
— |
|
|
|
|
| C. zyginae FM178171
|
1.4 |
3.8 |
1.4 |
2.6 |
— |
|
|
|
| C. guanche clade A
|
1.4 |
3.8 |
3.2 |
1.4 |
2.9 |
— |
|
|
| C. guanche clade B
|
1.4 |
3.8 |
3.2 |
2.0 |
2.9 |
0.6 |
— |
|
| C. guanche clade C
|
1.4 |
3.8 |
3.2 |
2.0 |
2.9 |
0.6 |
0.6 |
— |
|
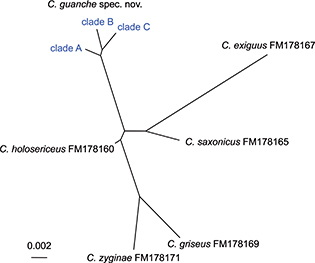 Fig 5.— Unrooted NJ-phenogram of genetic divergence of ITS2 genotypes. Scale bar indicates number of substitutions per amino acid
position. Fig 5.— Unrooted NJ-phenogram of genetic divergence of ITS2 genotypes. Scale bar indicates number of substitutions per amino acid
position.
Fig 5.— Fenograma NJ sin raíz de la divergencia genética de los genotipos del ITS2. La escala indica el número
de substituciones por cada posición aminoacídica.
|
|
|
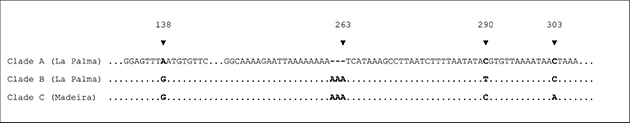 Fig. 6.— Excerpt of ITS2 alignment illustrating the intraspecific variability in C. guanche sp. nov. Fig. 6.— Excerpt of ITS2 alignment illustrating the intraspecific variability in C. guanche sp. nov.
Fig. 6.— Extracto del alineamiento del ITS2 mostrando la variabilidad intraespecífica de C. guanche sp. nov.
|
|
In four specimens of clade A (#2, #17, #20, #21), a second fragment of about 220bp amplified with somewhat lower signal strength.
This sequence is identical to ITS2 but exhibit a 151bp long deletion from alignment position 167 to 318, resulting in an interference
of 75bp in the chromatograms around positions 170 to 240. This shorter sequence was disregarded in the analyses performed
but might be an indication for intragenomic variation. An example of this shorter sequence was sent to ENA for specimen #20
(LT715963). Of clade B and C, all 15 sequences show a weak n-1 sequence from alignment position 263 onwards after a 11-fold
adenine repeat. In clade A, the homologous section consists of a 8-fold adenine repeat and did not pose any sequencing problems.
Although intragenomic variation cannot be ruled out here either, such artefacts may also result from a slippage of the enzyme
during PCR amplification. For the present analyses, the low signal of the underlying n-1 sequence was disregarded. Interestingly,
all C. guanche samples have an additional 20bp prior to the reverse primer (ACCTCAACTCATATGGGATT). In a previous study (Kehlmaier & Assmann, 2010), this motive emerged in only few samples of Chalarinae (FM212652, FM212653, FM212655, FM213119, LN879371), and, slightly
modified, also in one Pipunculinae used as outgroup (FM212650). As this oligonucleotide has not been detected in representatives
of the C. exiguus species group before and its origin is most likely attributed to intragenomic variation as well, it has been excluded from
the present analyses, yet included in ENA submissions.
The 5’ end of the mitochondrial cytochrome oxidase subunit I (COI) is the classical DNA barcoding fragment sequenced for a
multitude of taxa (Hebert et al., 2003). The aligned COI dataset for the Chalarus exiguus species group is 658bp long and consists of 40 specimens representing six species. Thirty specimens from La Palma (24 clade
A and 6 clade B) and five from Madeira (all clade C) yielded a DNA barcode with moderately to very good sequence quality.
The observed genetic divergence (p-distance) between the individual clades ranges from 1.2–3.1% between clades A and B, 1.2–1.8%
between clades A and C, and 0.6–1.8% between clades B and C. The maximum intraspecific variability is 0.8% in clade A, 1.2%
in clade B, and 0% in clade C. The minimum interspecific genetic distance between the species of the exiguus group ranges from 2.6% (C. guanche sp. nov. to C. griseus and C. saxonicus respectively) to 6.8% (C. exiguus to C. holosericeus) (Table 5). Interestingly, two specimens (#10, #21) with a clade A ITS2 genotype cluster with the specimens that have a clade B ITS2
genotype, providing evidence for hybridisation between both clades (Fig. 7).
Table 5.— Uncorrected minimum interspecific genetic distance in % of Chalarus exiguus species group for COI dataset. Roman numerals i)–xiii) are place holders for the individual haplotypes of C. guanche sp. nov. The individual clades of COI are colour coded.
Tabla 5.— Distancia genética interespecífica mínima no corregida
en tanto por ciento del grupo de especies de Chalarus exiguus para el conjunto de datos de la COI. Los numerales romanos i)-xiii) son marcadores para los haplotipos individuales de C. guanche sp. nov. Se han codificado con colores los clados individuales de COI.
| |
|
FM178136 |
FM178127 |
FM178120 |
FM178131 |
FM178122 |
i) |
ii) |
iii) |
iv) |
v) |
vi) |
vii) |
viii) |
ix) |
x) |
xi) |
xii) |
| ITS2 clade |
|
|
|
|
|
C |
B |
B |
B |
B |
B |
A |
A |
A |
A |
A |
A |
| C. holosericeus FM178136
|
|
— |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| C. saxonicus FM178127
|
|
4.0 |
— |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| C. exiguus FM178120
|
|
6.8 |
5.3 |
— |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| C. griseus FM178131
|
|
3.5 |
3.5 |
6.2 |
— |
|
|
|
|
|
|
|
|
|
|
|
|
|
| C. zyginae FM178122
|
|
4.2 |
4.8 |
6.1 |
4.4 |
— |
|
|
|
|
|
|
|
|
|
|
|
|
| i) M1–5 |
C |
3.0 |
2.6 |
5.5 |
2.6 |
3.7 |
— |
|
|
|
|
|
|
|
|
|
|
|
| ii) #3 |
B |
4.3 |
3.4 |
5.8 |
3.7 |
4.1 |
1.8 |
— |
|
|
|
|
|
|
|
|
|
|
| iii) #1 |
B |
3.8 |
3.2 |
5.8 |
3.3 |
4.2 |
1.5 |
1.1 |
— |
|
|
|
|
|
|
|
|
|
| iv) #8 |
B |
4.1 |
3.2 |
5.8 |
3.3 |
4.4 |
1.5 |
1.1 |
0.3 |
— |
|
|
|
|
|
|
|
|
| v) #7, 32 |
B |
4.1 |
3.1 |
5.8 |
3.4 |
4.2 |
1.5 |
0.0 |
0.8 |
0.8 |
— |
|
|
|
|
|
|
|
| vi) #36 |
B |
3.2 |
2.8 |
5.4 |
2.8 |
3.6 |
0.6 |
0.6 |
0.6 |
0.6 |
0.3 |
— |
|
|
|
|
|
|
| vii) #10 |
A |
3.5 |
2.9 |
5.3 |
2.9 |
3.8 |
1.1 |
0.8 |
0.0 |
0.2 |
0.6 |
0.3 |
— |
|
|
|
|
|
| viii) #21 |
A |
4.0 |
3.0 |
5.9 |
3.5 |
4.4 |
1.7 |
1.2 |
0.2 |
0.5 |
0.9 |
0.8 |
0.2 |
— |
|
|
|
|
| ix) #11, 18, 29, 38, 39, 41–43, 45–48 |
A |
3.3 |
3.2 |
5.8 |
3.0 |
4.0 |
1.2 |
2.4 |
2.1 |
2.1 |
2.1 |
1.2 |
1.7 |
2.3 |
— |
|
|
|
| x) #27, 40, 44 |
A |
3.5 |
3.0 |
5.8 |
3.2 |
4.0 |
1.2 |
2.5 |
2.1 |
2.1 |
2.1 |
1.2 |
1.7 |
2.3 |
0.2 |
— |
|
|
| xi) #2, 15, CK417 |
A |
3.8 |
3.3 |
5.9 |
3.5 |
4.2 |
1.4 |
2.6 |
2.3 |
2.3 |
2.3 |
1.4 |
1.8 |
2.4 |
0.5 |
0.2 |
— |
|
| xii) #12, 14, 19 |
A |
3.8 |
3.3 |
5.9 |
3.5 |
4.5 |
1.7 |
2.9 |
2.6 |
2.6 |
2.6 |
1.7 |
2.1 |
2.7 |
0.5 |
0.6 |
0.6 |
— |
| xiii) #20 |
A |
4.0 |
3.5 |
6.1 |
3.3 |
4.7 |
1.8 |
3.1 |
2.7 |
2.7 |
2.7 |
1.8 |
2.3 |
2.9 |
0.6 |
0.8 |
0.8 |
0.2 |
|
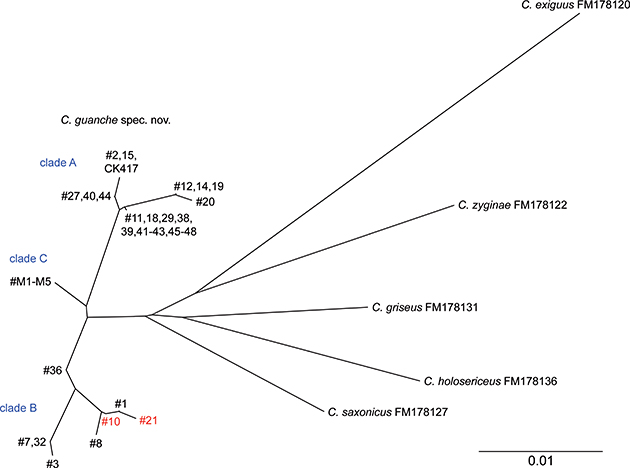 Fig. 7.— Unrooted NJ-phenogram of genetic divergence of COI haplotypes. Highlighted in red are the two specimens that indicate hybridisation
by sharing a clade A ITS2 genotype. Scale bar indicates number of substitutions per amino acid position. Fig. 7.— Unrooted NJ-phenogram of genetic divergence of COI haplotypes. Highlighted in red are the two specimens that indicate hybridisation
by sharing a clade A ITS2 genotype. Scale bar indicates number of substitutions per amino acid position.
Fig. 7.— Fenograma
NJ sin raíz de la divergencia genética de los haplotipos de la COI. Resaltados en rojo, los dos ejemplares que indican hibridación
al compartir un genotipo del ITS2 del clado A. La escala indica el número de sustituciones por posición aminoacídica.
|
|
DiscussionTOP
One of the great challenges of taxonomy is the identification and differentiation between closely allied species. Over the
past decades, and with the development of new taxonomic tools, integrative and iterative approaches have been advocated that
combine multiple lines of evidence, resulting in a multidimensional synthesis of known facts (Padial et al., 2010; Schlick-Steiner et al., 2010; Yeates et al., 2011). Here, the data at hand bring up the question whether the observed morphological and molecular heterogeneity in studied
Chalarus might justify the naming of additional species other than C. guanche sp. nov.
Morphological characters for distinguishing between species of Chalarus are scarce. Whereas male species diagnosis is almost exclusively based on the genitalia, females have to be identified via
an array of outer morphological features that can be hard to assess: shape of ovipositor, length of pulvilli, chaetotaxy of
legs, size of frontal facets and width of frons. However, deformation of the exoskeleton frequently takes place once the insect
is pinned and dried, especially if it was formerly preserved in a liquid. Bristles can break off, the pulvilli may shrivel,
the frons can experience a narrowing or the head may partly collapse or fall off. In addition, these artefacts meet with a
natural degree of morphological variability within a species. The 25 female specimens suitable for morphometric measurements
(Table 2) display a large morphological variability, as seen in the diameter of largest frontal facet (DFF), width of frons at level
of anterior/median ocellus (FMO), wing length and length of tergite 9. When the morphometric results are separated by clades
A and B (Table 3) only wing length and length of tergite 9 show a weak/marginal separation between the two lineages. All other morphological
traits and ratios do at least overlap slightly.
The results of the molecular analyses also display an ambiguous picture. On the one hand, haplotype divergence (maximum intraspecific
genetic distance) of COI between the specimens of C. guanche reaches 3.1%, and thus is higher than the minimum interspecific genetic distance between the species pairs C. guanche/C. saxonicus and C. guanche/C. holosericeus (2.6%) and almost as high as C. holosericeus/C. griseus and C. saxonicus/C. griseus (3.5%). On the other hand, genotype divergence between the three clades of C. guanche for the ITS2 dataset is 0.6%, and thus about half the smallest minimum interspecific genetic distance observed in this species
group (1.1% between C. holosericeus/C. saxonicus). Also, hybridisation between clades A and B could be detected in two cases.
A parenthesis has to be made in regard to the possible presence of intragenomic variability in ITS2. In eukaryotic cells,
ITS2, as part of the rDNA gene cluster, is arranged in multiple (typically several hundred) tandem repeats, which are known
to exhibit a certain degree of genetic variability. Several studies have investigated this phenomenon for ITS2 for different
taxa, e.g., bacteria (Stewart & Cavanaugh, 2007), fungi (Lindner & Banik, 2011; O’Donnell & Cigelnik, 1997), plants (Weitemier et al., 2015), corals (Odorico & Miller, 1997), sponges (Wörheide et al., 2004), crayfish (Harris & Crandall, 2000), molluscs (Hoy & Rodriguez, 2013), butterflies (Shapoval & Lukhtanov, 2015) and mosquitos (Bourke et al., 2013; Li & Wilkerson, 2007; Vesgueiro et al., 2011). The degree of intragenomic variation is mostly low due to concerted evolution (Brown et al., 1972). However, whenever concerted evolution is outrun by speciation, divergent paralogs, with up to 5% p-distance and more, have
been recorded in a single genome (e.g., Wörheide et al., 2004). Such paralogs even exceed interspecific variability (Li & Wilkerson, 2007) and ultimately lead to confound tree topology if included in phylogenetic analyses (Lindner & Banik, 2011). In the present study, the observed artefacts point towards this phenomenon, although a final proof can only be achieved
by cloning and the subsequent sequencing of multiple clones from a single organism.
In a nutshell, both loci support the validity of C. guanche sp. nov. as a separate taxon, but also leave room as to whether there are additional species present on Macaronesia. Extra insights
might be obtained through more intense sampling on all Macaronesian islands, especially in respect to the vertical distribution
and possible host associations of the individual clades. The former can currently be characterised by the presence of clade
B restricted to the two lower most localities (750–1,068 m), whereas clade A is distributed between 750–2,250 m. Knowledge
of the larval host species of the individual lineages of C. guanche might also be enlightening, and can be achieved by sampling and dissecting Typhlocybinae leafhoppers—the only known hosts
of Chalarus—in order to extract any larvae which can then be identified via DNA barcodes.
At present, the known Pipunculidae fauna of La Palma comprise the following seven species: Chalarus guanche, Dasydorylas setosus, Eudorylas fluviatilis, Tomosvaryella brachybasis, T. freidbergi, T. geniculata and T. parakuthyi.
AcknowledgementsTOP
We are much indebted to Dr. Michael von Tschirnhaus (Bielefeld) for making the material from Madeira in his collection (the
latter recently transferred to SDEI and in the care of Dr. Frank Menzel) available to us.
The first author is grateful for the financial support received from the SYNTHESYS Project http://www.synthesys.info/ financed by European Community Research Infrastructure Action under the FP6 ‘Structuring the European Research Area’ programme
to carry out part of the laboratory work at MNCN in Madrid.
The second author wants to thank the Direction and staff of the Park for their help during the two years collecting. He has
been partly supported by project CGL2015-66571-P (MINECO/FEDER) (Ministerio de Economía y Competitividad, Spain).
FOOTNOTESTOP
[1] | #44 MNCN_Ent 160692; #45 MNCN_Ent 160693; #46 MNCN_Ent 160694; # 47 MNCN_Ent 160691 |
[2] | #19 MNCN_Ent 160703; #20 MNCN_Ent 160704 |
[3] | #40 MNCN_Ent 160700; #41 MNCN_Ent 160701 |
[4] | #10 MNCN_Ent 160696; #11 MNCN_Ent 160697; #12 MNCN_Ent 160698; #13 MNCN_Ent 160699 |
[5] | #58 MNCN_Ent 202030; #59 MNCN_Ent 202031; #60 MNCN_Ent 202032; # 61 MNCN_Ent 202033 |
[6] | #52 MNCN_Ent 202037; #53 MNCN_Ent 202038 |
ReferencesTOP
| ○ |
Astrin, J. & Stüben, P., 2008. Phylogeny in cryptic weevils: molecules, morphology and new genera of western Palaearctic Cryptorhynchinae
(Coleoptera: Curculionidae). Invertebrate Systematics, 22(5): 503–522. https://doi.org/10.1071/IS07057 |
| ○ |
Beebe, N. W. & Saul, A., 1995. Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction-restriction fragment length polymorphism analysis. The American Journal of Tropical Medicine and Hygiene, 53: 478–481. https://doi.org/10.4269/ajtmh.1995.53.478 |
| ○ |
Bourke, B. P., Porangaba Oliveira, T., Suesdek, L., Sterlino Bergo, E. & Mureb Sallum, M. A., 2013. A multi-locus approach
to barcoding in the Anopheles strodei subgroup (Diptera: Culicidae). Parasites & Vectors, 6: 111, 16 pp. https://doi.org/10.1186/1756-3305-6-111 |
| ○ |
Brown, D. D., Wensink, P. C. & Jordan, E., 1972. Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. Journal of Molecular Biology, 63: 57–73.
|
| ○ |
De Meyer, M., 1995. The pipunculid flies of Israel and the Sinai (Insecta, Diptera, Pipunculidae). Spixiana, 18: 283–319.
|
| ○ |
De Meyer, M., Földvári, M. & Báez, M., 2001. The Pipunculidae (Diptera) fauna of the Canary Islands and Madeira. Bulletin et Annales de la Société Royale belge d’Entomologie, [2000], 136: 144–152.
|
| ○ |
Domingo-Quero, T., Alonso-Zarazaga, M.A., Sánchez-Ruiz, A., Araujo Armero, R., Navas Sánchez, A., Sánchez Moreno, S., García
Becerra, R., Nebreda, M., Sánchez Ruiz, M., Fontal-Cazalla, F. & Nieves-Aldrey, J.L., 2003. Inventariando la biodiversidad
en el Parque Nacional de la Caldera de Taburiente (La Palma, Islas Canarias, España): novedades científicas. Graellsia, 59(2–3): 45–68. https://doi.org/10.3989/graellsia.2003.v59.i2-3.235 |
| ○ |
Földvári, M. & De Meyer, M., 1999. Revision of Central and West European Tomosvaryella Aczél species (Diptera, Pipunculidae). Acta Zoologica Scientiarum Hungaricae, 45: 299–334.
|
| ○ |
Fregel, R., Pestano, J., Arnay, M., Cabrera, V. M., Larruga, J. M. & González, A. M., 2009. The maternal aborigine colonization
of La Palma (Canary Islands). European Journal of Human Genetics, 17(10): 1314–1324. https://doi.org/10.1038/ejhg.2009.46 |
| ○ |
Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.
Nucleic Acids Symposium Series, 41: 95–98.
|
| ○ |
Harris, D. J. & Crandall, K. A., 2000. Intragenomic variation within ITS1 and ITS2 of freshwater crayfishes (Decapoda: Cambaridae):
implications for phylogenetic and microsatellite studies. Molecular Biology and Evolution, 17(2): 284–291. https://doi.org/10.1093/oxfordjournals.molbev.a026308 |
| ○ |
Hebert P. D. N., Cywinska A., Ball S. L. & deWaard J. R., 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London. Series B, Biological Sciences, 270: 313–321. https://doi.org/10.1098/rspb.2002.2218 |
| ○ |
Hoy, M. S. & Rodriguez, R. J., 2013. Intragenomic sequence variation at the ITS1–ITS2 region and at the 18S and 28S nuclear
ribosomal DNA genes of the New Zealand mud snail, Potamopyrgus antipodarum (Hydrobiidae: Mollusca). Journal of Molluscan Studies, 79: 205–217. https://doi.org/10.1093/mollus/eyt016 |
| ○ |
Kehlmaier, C., 2005. Taxonomic revision of European Eudorylini. Verhandlungen des Naturwissenschaftlichen Vereins in Hamburg (Nova Folia), 41: 45–353.
|
| ○ |
Kehlmaier, C. & Assmann, T., 2008. The European species of Chalarus Walker, 1834 revisited (Diptera: Pipunculidae). Zootaxa, 1936: 1–39.
|
| ○ |
Kehlmaier, C. & Assmann, T., 2010. Molecular analysis meets morphology-based systematics—a synthetic approach for Chalarinae
(Insecta: Diptera: Pipunculidae). Systematic Entomology, 35: 181–195. https://doi.org/10.1111/j.1365-3113.2009.00500.x |
| ○ |
Li, C. & Wilkerson, R. C., 2007. Intragenomic rDNA ITS2 variation in the neotropical Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae). Journal of Heredity, 98(1): 51–59. https://doi.org/10.1093/jhered/esl037 |
| ○ |
Lindner, D. L. & Banik, M. T., 2011. Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and
inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia, 103(4): 731–740. https://doi.org/10.3852/10-331 |
| ○ |
O’Donnell, K. & Cigelnik, E., 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus
Fusarium are nonorthologous. Molecular Phylogenetics and Evolution, 7(1): 103–116.
|
| ○ |
Odorico, D. M. & Miller, D. J., 1997. Variation in the ribosomal internal transcribed spacers and 5.8s rDNA among five species
of Acropora (Cnidaria; Scleractinia): patterns of variation consistent with reticulate evolution. Molecular Biology and Evolution, 14(5): 465-473. https://doi.org/10.1093/oxfordjournals.molbev.a025783 |
| ○ |
Padial, J. M., Miralles, A., De la Riva, I. & Vences, M., 2010. The integrative future of taxonomy. Frontiers in Zoology, 2010:7, 16 pp. https://doi.org/10.1186/1742-9994-7-16 |
| ○ |
Rafael, J. A. & Skevington, J. H., 2010. Pipunculidae big-headed flies). In: B.V. Brown, A. Borkent, J.M. Cumming, D.M. Wood,
N.E. Woodley & M.A. Zumbado (eds.). Manual of Central American Diptera. Volume 2. NRC Research Press. Ottawa: 793–803.
|
| ○ |
Schlick-Steiner B. C., Steiner, F. M., Seifert, B., Stauffer, C., Christian, E. & Crozier, R. H., 2010. Integrative taxonomy:
a multisource approach to exploring biodiversity. Annual Review of Entomology 2010, 55: 421–438. https://doi.org/10.1146/annurev-ento-112408-085432 |
| ○ |
Shapoval, N. A. & Lukhtanov, V. A., 2015. Intragenomic variations of multicopy ITS2 marker in Agrodiaetus blue butterflies (Lepidoptera, Lycaenidae). Comparative Cytogenetics, 9(4): 483–497. https://doi.org/10.3897/CompCytogen.v9i4.5429 |
| ○ |
Stewart, F. J. & Cavanaugh, C. M., 2007. Intragenomic variation and evolution of the internal transcribed spacer of the rRNA
operon in Bacteria. Journal of Molecular Evolution, 65: 44–67. https://doi.org/10.1007/s00239-006-0235-3 |
| ○ |
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S., 2013. MEGA6: Molecular Evolutionary Genetics Analysis version
6.0. Molecular Biology and Evolution, 30: 2725–2729. https://doi.org/10.1093/molbev/mst197 |
| ○ |
Vesgueiro, F. T., Demari-Silva, B., Malafronte, R. dos S., Sallum, M. A. M. & Marrelli, M. T., 2011. Intragenomic variation
in the second internal transcribed spacer of the ribosomal DNA of species of the genera Culex and Lutzia (Diptera: Culicidae). Memórias do Instituto Oswaldo Cruz, 106(1): 1–8. https://doi.org/10.1590/S0074-02762011000100001 |
| ○ |
Weitemier, K., Straub, S. C. K., Fishbein, M. & Liston, A., 2015. Intragenomic polymorphisms among high-copy loci: a genus-wide
study of nuclear ribosomal DNA in Asclepias (Apocynaceae). PeerJ, 3: e718, 23 pp. https://doi.org/10.7717/peerj.718 |
| ○ |
Wörheide, G., Nichols, S. A. & Goldberg, J., 2004. Intragenomic variation of the rDNA internal transcribed spacers in sponges
(Phylum Porifera): implications for phylogenetic studies. Molecular Phylogenetics and Evolution, 33: 816–830. https://doi.org/10.1016/j.ympev.2004.07.005 |
| ○ |
Yeates, D. K., Seago, A., Nelson, L., Cameron, S. L., Joseph, L. & Trueman, J. W. H., 2011. Integrative taxonomy, or iterative
taxonomy? Systematic Entomology, 36(2): 209–217. https://doi.org/10.1111/j.1365-3113.2010.00558.x |
Appendix 1.— Specimen, locality and collecting data of studied material of C. guanche sp. nov., including GenBank accession numbers and clade affiliation. Abbreviations: MT, Malaise trap; YT, yellow pan trap. *Specimen
misplaced. **Only genitalia left.
Apéndice 1.— Ejemplar, localidad y datos de colecta del material estudiado de C. guanche sp. nov., incluyendo los números de registro en GenBank y la afiliación a clados. Abreviaturas: MT, trampa Malaise; YT, trampa Moericke
amarilla. *Ejemplar extraviado. **Sólo se conserva la genitalia.TOP
| sex |
date |
trap |
specimen no. |
ITS2 clade |
ITS2 |
COI |
| Spain, Canary Islands, La Palma, Playa del Río Taburiente. |
|
|
|
|
|
|
| ♀ |
6.III.2000 |
MT |
#25 |
A |
LT715939 |
— |
| ♂ |
17.IV.2000 |
MT |
#55 |
— |
— |
— |
| ♀ |
17.IV.2000 |
MT |
#23 |
B |
LT715940 |
— |
| ♀ |
15.V.2000 |
MT |
#22 |
— |
— |
— |
| ♂ |
19.VI.2000 |
YT |
#57 |
— |
— |
— |
| ♂ |
3.VIII.2000 |
YT |
#58 |
— |
— |
— |
| ♂ |
3.VIII.2000 |
YT |
#59 |
— |
— |
— |
| ♂ |
3.VIII.2000 |
YT |
#60 |
— |
— |
— |
| ♂ |
3.VIII.2000 |
YT |
#61 |
— |
— |
— |
| ♀ |
3.VIII.2000 |
YT |
#24 |
— |
— |
— |
| ♀ |
7.VIII.2000 |
MT |
#26 |
B |
LT715941 |
— |
| ♂ |
18.X.1999 |
MT |
#56 |
— |
— |
— |
| Spain, Canary Islands, La Palma, Barranco de las Traves. |
|
|
|
|
|
|
| ♀ |
8.V.2000 |
MT |
#8 |
B |
LT715942 |
LT715904 |
| ♀ |
30.V.2000 |
MT |
#6 |
B |
LT715943 |
— |
| ♂ |
5.VI.2000 |
MT |
#35 |
— |
— |
— |
| ♂ |
13.VI.2000 |
YT |
#29 |
A |
LT715944 |
LT715905 |
| ♂ |
13.VI.2000 |
MT |
#30 |
— |
— |
— |
| ♀ |
19.VI.2000 |
MT |
#1 |
B |
LT715945 |
LT715906 |
| ♂ |
27.VI.2000 |
MT |
#33 |
— |
— |
— |
| ♂ |
4.VII.2000 |
MT |
#34 |
B |
LT715946 |
— |
| ♂ |
10.VII.2000 |
YT |
#28 |
— |
— |
— |
| ♂ |
24.VII.2000 |
YT |
#32 |
B |
LT715947 |
LT715907 |
| ♀ |
24.VII.2000 |
MT |
#3 |
B |
LT715948 |
LT715908 |
| ♀ |
3.VIII.2000 |
YT |
#4 |
— |
— |
— |
| ♀ |
3.VIII.2000 |
YT |
#5 |
— |
— |
— |
| ♂ |
7.VIII.2000 |
MT |
#36 |
B |
LT715949 |
LT715909 |
| ♀ |
7.VIII.2000 |
MT |
#7 |
B |
LT715950 |
LT715910 |
| ♀ |
22.VIII.2000 |
MT |
#2 |
A |
LT715951 |
LT715911 |
| ♂ |
18.IX.2000 |
YT |
#31 |
— |
— |
— |
| Spain, Canary Islands, La Palma, Lomo de las Chozas. |
|
|
|
|
|
|
| ♂ |
5.I.2000 |
MT |
#51 |
— |
— |
— |
| ♀ |
8.III.2000 |
MT |
#21 |
A |
LT715952 |
LT715912 |
| ♂ |
22.III.2000 |
YT |
#48 |
A |
LT715953 |
LT715913 |
| ♂ |
26.IV.2000 |
MT |
#49** |
— |
— |
— |
| ♀ |
26.IV.2000 |
MT |
#15 |
A |
LT715954 |
LT715914 |
| ♀ |
26.IV.2000 |
MT |
#16* |
— |
— |
— |
| ♂ |
1.VI.2000 |
MT |
#37** |
— |
— |
— |
| ♀ |
1.VI.2000 |
MT |
#9* |
— |
— |
— |
| ♂ |
21.VI.2000 |
MT |
#43 |
A |
LT715955 |
LT715915 |
| ♂ |
21.VI.2000 |
MT |
#44 |
A |
LT715956 |
LT715916 |
| ♂ |
21.VI.2000 |
MT |
#45 |
A |
LT715957 |
LT715917 |
| ♂ |
21.VI.2000 |
MT |
#46 |
A |
LT715958 |
LT715918 |
| ♂ |
21.VI.2000 |
MT |
#47 |
A |
LT715959 |
LT715919 |
| ♀ |
21.VI.2000 |
MT |
#14 |
A |
LT715960 |
LT715920 |
| ♂ |
6.VII.2000 |
MT |
#52 |
— |
— |
— |
| ♂ |
6.VII.2000 |
MT |
#53 |
— |
— |
— |
| ♂ |
6.VII.2000 |
MT |
#54 |
— |
— |
— |
| ♀ |
6.VII.2000 |
MT |
#19 |
A |
LT715961 |
LT715921 |
| ♀ |
6.VII.2000 |
MT |
#20 |
A |
LT715962 |
LT715922 |
| ♀ |
6.VII.2000 |
MT |
#20 |
A short |
LT715963 |
|
| ♂ |
19.VII.2000 |
MT |
#40 |
A |
LT715964 |
LT715923 |
| ♂ |
19.VII.2000 |
MT |
#41 |
A |
LT715965 |
LT715924 |
| ♂ |
19.VII.2000 |
MT |
#42 |
A |
LT715966 |
LT715925 |
| ♀ |
19.VII.2000 |
MT |
#10 |
A |
LT715967 |
LT715926 |
| ♀ |
19.VII.2000 |
MT |
#11 |
A |
LT715968 |
LT715927 |
| ♀ |
19.VII.2000 |
MT |
#12 |
A |
LT715969 |
LT715928 |
| ♀ |
19.VII.2000 |
MT |
#13 |
A |
LT715970 |
— |
| ♂ |
26.VII.2000 |
MT |
#50 |
— |
— |
— |
| ♀ |
26.VII.2000 |
MT |
#17 |
A |
LT715971 |
— |
| ♂ |
16.VIII.2000 |
MT |
#38 |
A |
LT715972 |
LT715929 |
| ♂ |
16.VIII.2000 |
MT |
#39 |
A |
LT715973 |
LT715930 |
| ♀ |
6.IX.2000 |
MT |
#18 |
A |
LT715974 |
LT715931 |
| Spain, Canary Islands, La Palma, Roque de los Muchachos. |
|
|
|
|
|
|
| ♂ |
27.VII.2000 |
YT |
#62 |
— |
— |
— |
| ♀ |
25.VIII.2000 |
MT |
#27 |
A |
LT715975 |
LT715932 |
| Spain, Canary Islands, La Palma, south of Barlovento, laurisilva, 28°39’N 17°52’W, 800 m, leg. et coll. C. Kehlmaier |
|
|
|
|
|
|
| ♂ |
29.X.2002 |
net |
DNA CK145 |
— |
— |
— |
| ♂ |
29.X.2002 |
net |
DNA CK146 |
— |
— |
— |
| ♂ |
29.X.2002 |
net |
DNA CK417 |
B |
LT715976 |
LT715933 |
| ♀ |
29.X.2002 |
net |
|
— |
— |
— |
| Portugal, Madeira, path between Boca de Encumeada and Boca dos Corgos, S of Ribeiro do Póco, 1 km W of Fenda do Ferreiro,
moist steep slope, 1,200–1,250 m, X572, leg. W. Barkemeyer, coll. SDEI
|
|
|
|
|
|
|
| ♂ |
30.V.1987 |
|
M1 |
C |
LT715977 |
LT715934 |
| Portugal, Madeira, Fanal, laurel on pasture, 12.IX.1986, L1583, leg. P. Ohm, coll. SDEI |
|
|
|
|
|
|
| ♂ |
12.IX.1986 |
|
M2 |
C |
LT715978 |
LT715935 |
| ♂ |
12.IX.1986 |
|
M3 |
C |
LT715979 |
LT715936 |
| ♂ |
12.IX.1986 |
|
M4 |
C |
LT715980 |
LT715937 |
| Portugal, Madeira, Pico Facho, near Machico, herbaceous vegetation, 300 m, L1578, leg. P. Ohm, coll. SDEI |
|
|
|
|
|
|
| ♂ |
19.IX.1986 |
|
M5 |
C |
LT715981 |
LT715938 |
Fig. 1.— Male genitalia of C. guanche sp. nov. in lateral view. Abbreviations: lower ej.d., lower ejaculatory ductuli; mtdp, membranous tip of distiphallus; php, phallic
processes; phs, phallic shaft; tdp, tip of distiphallus; upper ej.d., upper ejaculatory duct. Scale bar: 0.1 mm.